Predicting CTCF-mediated chromatin loops using CTCF-MP
Ruochi Zhang, Yuchuan Wang, Yang Yang, Yang Zhang, Jian Ma.
Published in ISMB 2018, Bioinformatics
Abstract
Motivation
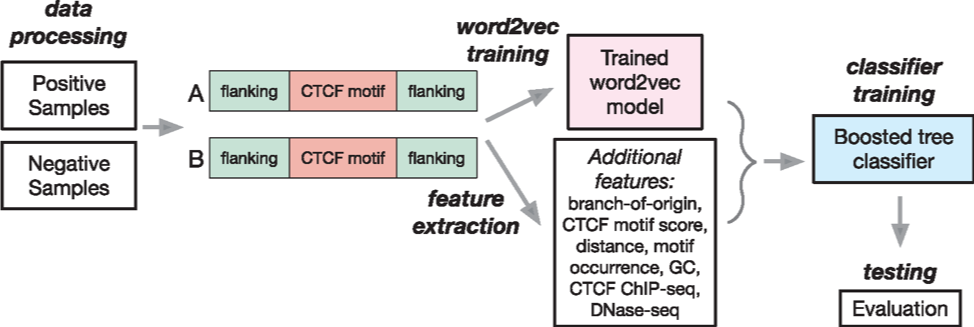
The three dimensional organization of chromosomes within the cell nucleus is highly regulated. It is known that CCCTC-binding factor (CTCF) is an important architectural protein to mediate long-range chromatin loops. Recent studies have shown that the majority of CTCF binding motif pairs at chromatin loop anchor regions are in convergent orientation. However, it remains unknown whether the genomic context at the sequence level can determine if a convergent CTCF motif pair is able to form a chromatin loop.
Results
In this article, we directly ask whether and what sequence-based features (other than the motif itself) may be important to establish CTCF-mediated chromatin loops. We found that motif conservation measured by ‘branch-of-origin’ that accounts for motif turn-over in evolution is an important feature. We developed a new machine learning algorithm called CTCF-MP based on word2vec to demonstrate that sequence-based features alone have the capability to predict if a pair of convergent CTCF motifs would form a loop. Together with functional genomic signals from CTCF ChIP-seq and DNase-seq, CTCF-MP is able to make highly accurate predictions on whether a convergent CTCF motif pair would form a loop in a single cell type and also across different cell types. Our work represents an important step further to understand the sequence determinants that may guide the formation of complex chromatin architectures.