Probing multi-way chromatin interaction with hypergraph representation learning
Ruochi Zhang, Jian Ma.
Published in RECOMB 2020, Cell Systems
Abstract
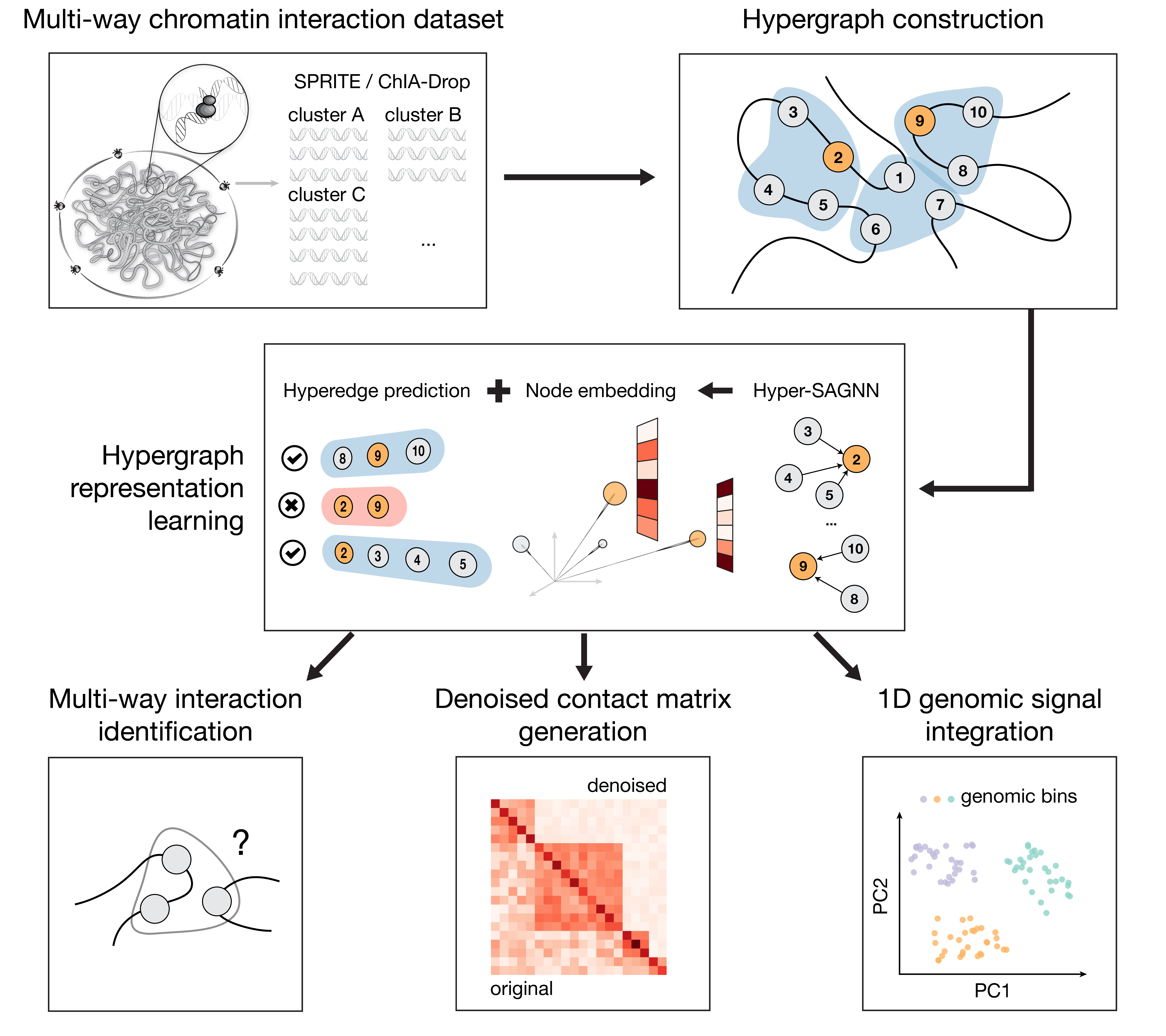
Advances in high-throughput mapping of 3D genome organization have enabled genome-wide characterization of chromatin interactions. However, proximity ligation based mapping approaches for pairwise chromatin interaction such as Hi-C cannot capture multi-way interactions, which are informative to delineate higher-order genome organization and gene regulation mechanisms at single-nucleus resolution. The very recent development of ligation-free chromatin interaction mapping methods such as SPRITE and ChIA-Drop has offered new opportunities to uncover simultaneous interactions involving multiple genomic loci within the same nuclei. Unfortunately, methods for analyzing multi-way chromatin interaction data are significantly underexplored. Here we develop a new computational method, called MATCHA, based on hypergraph representation learning where multi-way chromatin interactions are represented as hyperedges. Applications to SPRITE and ChIA-Drop data suggest that MATCHA is effective to denoise the data and make \textit{de novo} predictions of multi-way chromatin interactions, reducing the potential false positives and false negatives from the original data. We also show that MATCHA is able to distinguish between multi-way interaction in a single nucleus and combination of pairwise interactions in a cell population. In addition, the embeddings from MATCHA reflect 3D genome spatial localization and function. MATCHA provides a promising framework to significantly improve the analysis of multi-way chromatin interaction data and has the potential to offer unique insights into higher-order chromosome organization and function.